Spin column purification of nucleic acids - the biochemistry of how they work
ฝัง
- เผยแพร่เมื่อ 4 ต.ค. 2024
- If you’re looking for a spin class, have you tried out spin column DNA/RNA purification? I hope you don’t mind if my PCR products bind? Biochemists win when we take them for a spin! “Spin Columns” are a quick & easy way to purify pieces of DNA (or RNA) - just make sure you get the right kit! These columns are a form of “solid-phase extraction” where you bind the nucleic acid to a silica gel membrane, wash off the other stuff, then “un-bind” pure DNA or RNA. There are lots of different versions & kits & they work really similar so, although I’m going to be talking about PCR PURIFICATION (aka “clean-up”) the basic principles apply to other situations
full version (text old, video new): bit.ly/spincolumns
Let’s look at the players.
The membrane in silica-based. silica (amorphous silicon dioxide, SiO₂) can have a lot of modifications & the composition of these membranes are proprietary, so I don’t know exactly what their surfaces actually look like at the molecular level, but silica has hydroxyl (-OH) groups that at low pH (acidic, where there are lots of free protons (H⁺) floating around) will be protonated (-OH) but at higher pHs (where there are less free protons) will be deprotonated & thus negative (-O⁻). In addition to this group, silica also offers up some additional opportunities for hydrogen bonding & even hydrophobic interactions.
& the DNA? DNA’s a polymer (chain of similar repeating units) of nucleotides (DNA letters). These have a generic sugar-phosphate backbone they link through (same-strand bonds) & unique nitrogenous bases (A, T, G, or C) that stick out & form the basis of between-strand base pairing.
The phosphate in that sugar phosphate backbone is negatively charged, & it’s really happy being negatively charged, so even at pretty low pH it’s gonna stay that way. “Normally” the nitrogenous bases are neutral. BUT, as the name suggests, they are (weak) bases, meaning that if the pH is low enough (meaning there are lots of H⁺ around) they will pick one up & this makes them + charged. So, at lower pH, the overall charge of DNA decreases, so it becomes less water-soluble.
This helps us separate DNA from RNA in liquid-phase RNA extraction. At a low pH, DNA prefers the organic phenol-chloroform phase, whereas RNA’s extra -OH keeps it in the aqueous phase. bit.ly/rnaextra...
Here, at a low pH, DNA doesn’t have an “organic phase” - instead, it binds (adsorbs to) the silica membrane. Note: I’m not sure if the pH is low enough here to change those bases, but there’s another important function of the pH.
At a low pH, the sometimes OH, sometimes O groups of the silica are more likely to be in the -OH form, so there’s less negative charge that could repel the negatively charged DNA backbone.
It’s not just the low pH that’s important. You also need salts. CHAOTROPIC SALTS like guanidinium hydrochloride bring “chaos” to water → they disrupt water’s bonding networks, “loosening up” the water coat surrounding the DNA so the DNA can seek out & bind to the silica instead. When it does so, you get an entropic benefit because it frees up water molecules that were stuck so they can move around more, thus increasing entropy (disorder)more here: bit.ly/ionicstr...
There are different ways it can bind, & the actual situations probably a combination of lots of them. The exposed bases could form direct H-bonds or hydrophobic interactions with the silica; Cations from the salts could form “bridges” between the backbone & the silica; etc.
The more interactions, the stronger the binding. The smaller DNA pieces (primers) don’t offer enough binding opportunities to stick well, so they flow right on through with the other stuff
So we start by mixing our (completed) PCR reaction with a solution containing guanidinium hydrochloride (the chaotropic salt to loosen the water shells) &, to help get the DNA to precipitate (come out of solution), ISOPROPANOL (aka isopropyl alcohol, aka propan-2-ol, etc.). This lowers the dielectric constant → reduces electrostatic shielding so that the salts, DNA, & silica can find each other. & it “dilutes” the water, so there’s less water available to bind the DNA, so the DNA “dehydrates” & really sticks on tight to that silica so it doesn’t come loose when you do the washes. Finished in comments






![เปิดบ้าน พี่ตูน Bodyslam หลังใหม่คนแรก ที่ภูเก็ต l [Nickynachat]](http://i.ytimg.com/vi/flFwcmyDQSk/mqdefault.jpg)

If you’re using a QIAquick PCR purification kit, this is buffer PB. (This isn’t a paid ad for Qiagen or anything, it’s just what our (and many) labs use).
The kit also includes a pH indicator you can (& should) put in! (unless you’re gonna be using the DNA for a super-sensitive microarray) - you want to make sure that the pH is low enough (below about 7.5). If the solution is orangey-purpley, it’s too high & the DNA won’t bind well. Don’t worry, just add a little acid (sodium acetate, pH 5 does the trick) → should turn yellow
Now you need to do the actual binding part.
The silica gel membrane is held in a microspin column you can transfer between tubes to collect the flow-through & you pipet your sample into the “cuppy” part above the membrane. Then you centrifuge it (spin it really fast) to pull the liquid through. (alternatively, if you have a lot of samples to do at once, it can be quicker to use a vacuum manifold which sucks it through (though you’ll still have to switch to spin for the elution because it sucks it through into the waste…)).
The bigger pieces of DNA (greater thann 100bp) will (hopefully) bind the membrane, but all the other stuff (primers, denatured DNA Pol, salts, etc.) will flow on through & you can toss it (the liquid, not the column!)
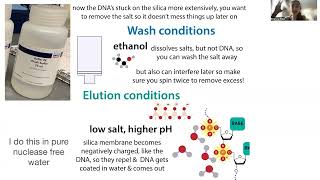
Now comes the wash. Now that you’ve gotten DNA stuck on there tight, you want to wash off any lingering extra salts. You do this using a buffer containing ethanol (buffer PE in Qiagen’s kit). just add the liquid the same way you added your sample, spin it through & toss it out.
DNA’s not soluble in ethanol, but salts are, so the extra salts are removed. It’s really important that you remove that guanidinium chloride because, while you wanted to denature the DNA Pol, you don’t want it to stick around & interfere with any reactions you do with the DNA later on. Finished in comments
After the wash & toss, spin it again to give any residual ethanol another chance to get pulled through. It’s really important to remove the ethanol or the DNA will have problems dissolving &/or when you go to load samples into an agarose gel (more here:bit.ly/2Z6vmeR) they’ll float up out of the well cuz ethanol’s less dense than the buffer.
Now that you’ve gotten the DNA clean, you need to get it to unstick from the silica. To do this, you need to reverse the conditions that got it to stick. We get it to stick with high salt, low pH → we get it to unstick with low salt, high pH.
The Qiagen kit comes with an elution buffer (buffer EB - which is 10 mM Tris·Cl, pH 8.5) (Tris is a buffering agent) you can use or you can use plain old (but really pure, nuclease-free) water. If you use water, give it a minute to fully dissolve the DNA before you spin it through.
Speaking of which → once you redissolve it, you spin it through into a NEW TUBE - this one you want to keep!
Then, take your spun-through, pure DNA over to the spectrometer like a NanoDrop & check out its absorbance spectrum (it should have a 260/280 ratio of ~1.8). More on that here: bit.ly/dnauvbeer
more practical tips for using these columns: th-cam.com/video/MBnuae5aWg4/w-d-xo.html
more about all sorts of things: #365DaysOfScience All (with topics listed) 👉 bit.ly/2OllAB0 or search blog: thebumblingbiochemist.com
Genius! It sounds like a product concluding many basic knowledge
Thank you for the great video and enthusiasm.
Glad you liked it!
Was very useful. 10/10 spin. Subbed
Great to hear it was helpful!
@@thebumblingbiochemist Have u made any videos on gentra kit method (DNA extraction) or anionic detergents?
not familiar with the gentra kit sorry - a bit on detergents here: th-cam.com/video/kV3tPr7ZT4o/w-d-xo.html
@@thebumblingbiochemist That's good enough. Thanks again. Off topic but long did it take you to acquire all this knowledge and how do you remember all this stuff
college, grad school, and still always learning (and Googling to refresh my memory and look up what I don't know)
Have you made any videos about RNA extraction by PureLink® RNA Mini Kit
Not that kit specifically no but the principles of them all are basically the same. Good luck!
I haven't get the diff between DNA , RNA extraction since they are the same principle why didnt they messed up together , can you explain pls
I am using spin column for rna extraction
A260/280 is 1.95 but the concentration is 21.2 ng/ul ,can you give me any suggestion to increase the yield.
If you want higher yield you may need to start with more sample. But you can try letting the elution buffer sit on the membrane for a minute before spinning. And you can increase the concentration by eluting in less volume.
so is spin column negatively or positivley charged 😃?
neutral at low pH, negative at higher pH