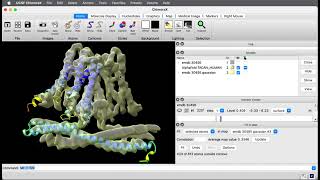
Run AlphaFold in ChimeraX for cryoEM model building
ฝัง
- เผยแพร่เมื่อ 20 ส.ค. 2024
- We show how to run AlphaFold from ChimeraX to predict a protein structure from its sequence to produce an initial model for building into a 3 Angstrom resolution cryoEM map. We look at a protein that transports omega-3 fatty acids across the blood-brain barrier, EMDB 23883. Requires ChimeraX 1.3 or a daily build newer than September 2021.





![[TH] 2024 PMSL SEA W1D5 | Fall | เก็บแต้มของจริง คือวันนี้](http://i.ytimg.com/vi/EciIAlCXg5k/mqdefault.jpg)


Hello, It is wonderful information you gave thank you. Could you please also provide some info or video on how to perform this same funtion if we have multi-domian herto protein structure with 2 or more subunits? will the same protocol can be applicable in that case or we need to apply some different strategy?
AlphaFold works on a single protein sequence. I have heard people try to put a linker between two proteins to make it a single sequence and then run AlphaFold with mixed results. I would not expect that to work well because the multiple sequence alignment AlphaFold produces to predict the long range contacts won't include sequences spanning the linker so I would not think it would provide any info about inter-protein contacts. Also AlphaFold crashes on longer sequences running out of GPU memory. The EBI AlphaFold database limited sequences to 1400 amino acids and that used the memory from 4 GPUs. The ChimeraX AlphaFold prediction can only use one GPU and seems limited to about 800 amino acids.
Very nice!