Molecular Docking; Protein-Ligand Docking In Vina (Part-1) How to Prepare Receptor File
ฝัง
- เผยแพร่เมื่อ 5 ก.พ. 2025
- Our course links on Udemy:Learn Docking and MD Simulation
www.udemy.com/...
Learn Bioinformatics from Scratch (Theory and Practical)
www.udemy.com/...
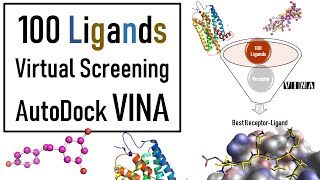
If are you interested to perform Molecular docking. Docking using Vina then you are right place? In this tutorial (Part 1) we will talk about that how you can prepare the Receptor files for Docking in Vina. Please stay with us. We will bring the next part soon.








this is very explanatory. how do i get the part 2
Hello sir..
I'm doing targeted docking . Is it necessary that the ligand should be in the grid box?
Yes it is important
@@bioinfoxpert ok sir
thank you 🙏
I have some more questions... Please reply if possible....
In the final result of autodock two types of RMSD are given. What are these two?
which RMSD value we should quote when we want to report the result for publication?
why does I have high value of reference RMSD? I though reference RMSD are supposed to be
How install docking molecules
Sir, Here do we need to minimize the energy?
yes if possible
Thank you! Part 2?
Soon
How can we make the protein and ligand complex after docking? Please reply
just open both in pymol and save them as single molecule.
part 2